Past Research
Allen Institute for Neural Dynamics
OpenScope Databook
Motivation for Project
My Role:
Built an end-to-end machine learning workflow to model mouse visual cortex activity and relate neural dynamics to visual stimuli.
Developed pipeline for neuron classification and spike sorting.
Reviewed and edited Databook notebooks for clarity and consistency, strengthening explanations, captions, and step-by-step guidance to make workflows accessible across neuroscience subfields and skill levels.
Other Credits (main contributors): Carter Peene, Jerome Lecoq, Ahad Bawney
GitHub: https://alleninstitute.github.io/openscope_databook/intro.html
Project Status: Published - click here
Reproducibility and validation are major challenges in neuroscience. Multimodal analyses and visualizations are complex and results are often difficult to replicate without accessible, open-source pipelines. The OpenScope Databook addresses this by bringing together the four essentials of reproducible research: accessible data, accessible computational resources, a reproducible environment, and clear usage documentation. We designed this project as a globally accessible resource that teaches scientists, educators, and students how to utilize computational and machine learning methods to analyze, visualize, and reproduce neural data analysis with open-source data. The notebooks can be run through Binder, Thebe, DANDIHub, or locally. Our goal is to build a framework and culture that prioritizes findings that are valid, transparent, and easy to reproduce across projects and datasets.
Technical Explanation of My Contributions:
1) Using CEBRA-Time to Identify Latent Embedding
click here to see my code
This notebook demonstrates how to use the CEBRA-Time algorithm, turning mouse visual-cortex recordings into a compact 3D “map” of neural activity during passive viewing of movies. This makes complex patterns easier to see and compare. The goal is to learn an embedding that stays consistent across repeated presentations of the same stimulus, revealing a shared neural representation of the movie that reappears across repeats.
Wrote the runnable walkthrough that takes a reader from “what is CEBRA-Time?” to a complete example analysis on open-source datasets, including setup steps so it works in shared compute environments like
Built the data-loading pipeline that pulls NWB file (adptable for any file) from DANDI and prepares it for analysis in a repeatable way.
Decoded the stimulus timeline (what the mouse was seeing, and when) directly from the NWB stimulus objects, so the notebook can label the data without manual formatting.
Solved the time-alignment problem by matching every neural measurement to the correct movie frame/stimulus type, handling edge cases where timestamps occur outside stimulus presentation, and trimming arrays so everything lines up cleanly.
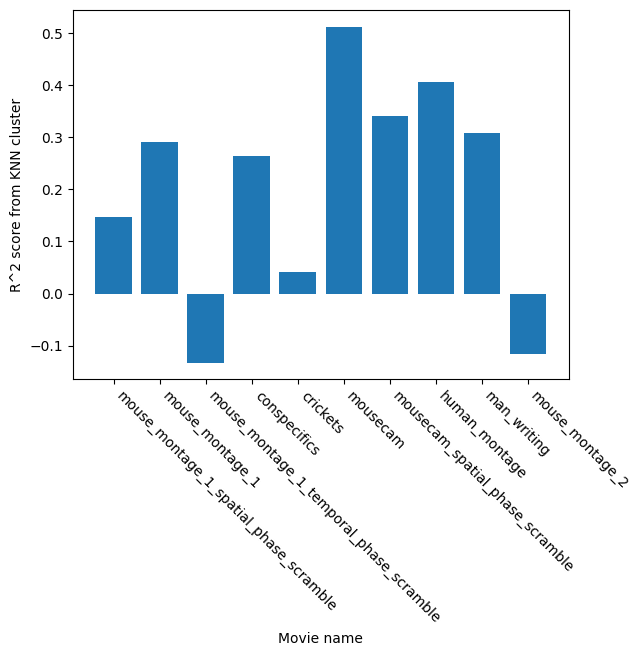
Implemented model training + evaluation outputs: ran a parameter search, trained and saved the final model, plotted training quality, generated intuitive 3D visualizations split by stimulus and repeat, and added a simple quantitative check to see how consistently “movie time” is represented across repeats.
2) Classifying Neuronal Types
click here to see my code
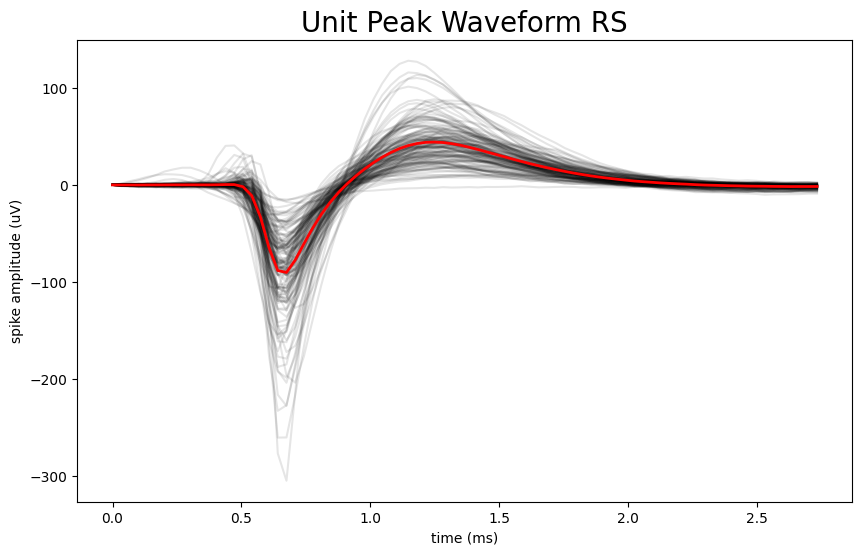
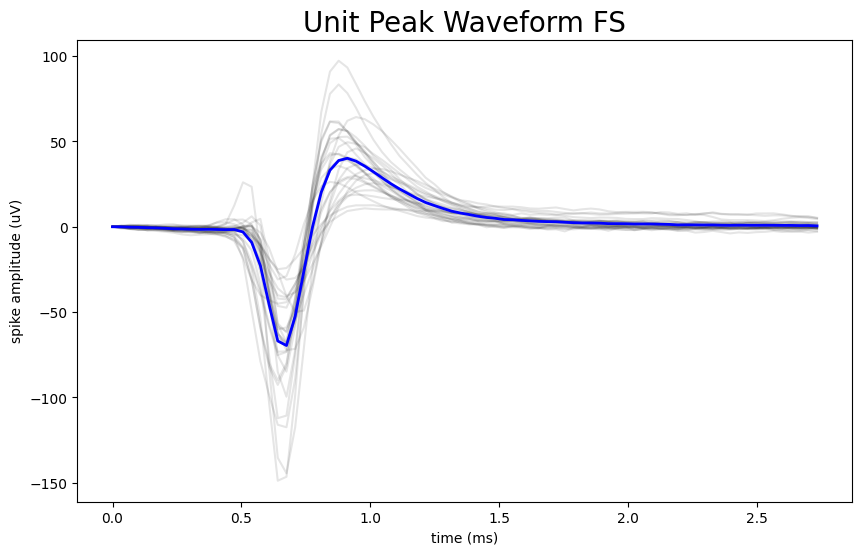
This notebook shows how to separate two common neuron types, fast-spiking and regular-spiking, using simple, interpretable features from each neuron’s recorded electrical waveform. It then visualizes what makes the groups different (waveform shape and response patterns) using Allen Institute OpenScope data, while keeping the workflow easy to adapt to other datasets.
Built a start-to-finish example that loads a public Allen Institute electrophysiology dataset (NWB) directly from DANDI so anyone can reproduce the analysis without manual file handling.
Wrote the unit-selection and quality-filtering logic to focus the analysis on reliable recordings from visual cortex (and made it easy to swap criteria for other datasets).
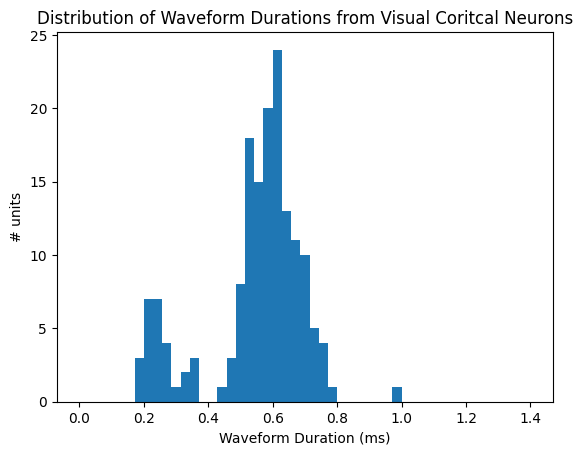
Created the classification workflow by measuring spike “width” (waveform duration) and using its natural two-peak distribution to separate fast-spiking vs regular-spiking units with a clear threshold.
Implemented the data alignment needed to connect neuron waveforms to the correct recording channel, bridging information stored across different NWB tables.
Produced the visual comparisons that make the result intuitive: a histogram of waveform durations and overlay plots showing typical waveform shapes for each neuron group.
—>
—>
Cluster Predictability Across Movie Stimuli