Current Research Projects
Dynamical Inference Lab
Brain Organoid Neural Dynamics
Motivation for Project
My Role:
Research Engineer for Brain Organoid Electrophysiology and Dynamics
Data architecture: implementing a multimodal DataJoint database schema for experiments, metadata, data validation, and preprocessing
ML engineering: utilized and tuned dynamical models and embeddings of neural activity, visualizations of results
Other Credits: Steffen Schneider, Silviu-Vasile Bodea, Lucas Mair, Zoe Zhuoya Wang, Rodrigo Gonzales
GitHub: unreleased repository
Project Status: in progress
Skills Used In Project (TLDR):
multimodal neural data analysis, database design and end-to-end data pipelines, signal processing (filtering, artifact rejection, common-average referencing), machine learning for neural dynamics (time-delay embeddings, DCL), dimensionality reduction and modeling (PCA, UMAP, regression analysis, ARHMM), Python (NumPy, SciPy, pandas, Matplotlib, PyTorch), HPC workflows (cluster jobs, resource configuration, fail-safes), data integrity and synchronization, visualization of data, biometric and physiological sensing workflows (electrophysiology, calcium imaging)
This project explores how human brain organoids develop, organize, and respond to stimulation through their electrical activity. A brain organoid is a collection of stem-cell-derived “mini-brain” that self-organizes into early neural tissue and forms developing networks capable of producing spontaneous electrical activity. A collaborating lab recorded activity from human brain organoids using multielectrode arrays across approximately 40 days. By systematically characterizing both spontaneous and evoked activity, we aim to understand how these networks develop and become structured, how their dynamics change across time, and what kinds of input-output behaviors they can support in a research setting. A long-term goal of the project is to move from offline analysis to real-time interaction. By training models that approximate organoid dynamics (“digital twins” of the living networks), we ultimately aim to build closed-loop systems where stimulation is adapted in real time to ongoing neural activity. This would turn brain organoids into active research tools in human-machine dialogues, opening new ways to probe, shape, and understand neural systems.
Technical Explanation of Personal Contributions:
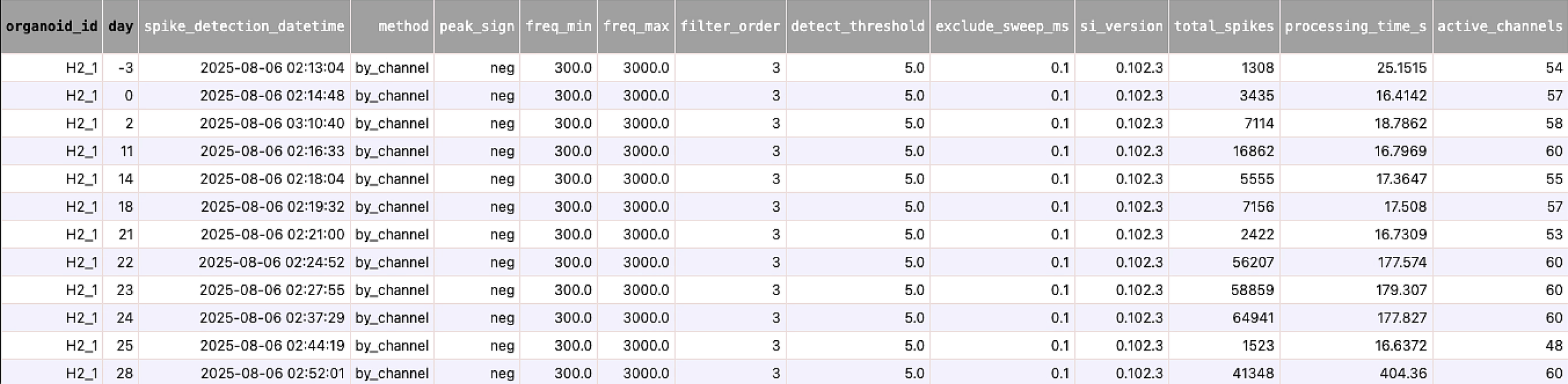
Data Infrastructure – Designed a DataJoint database and automated pipelines that take multi-day, multi-organoid MEA recordings from raw HDF5 files to analysis-ready tables (spikes, embeddings, QC), built and documented a command-line interface (CLI) for the database and analysis pipelines to make it easy to querying data and executing analysis accessible to all collaborators who are not familiar with the codebase
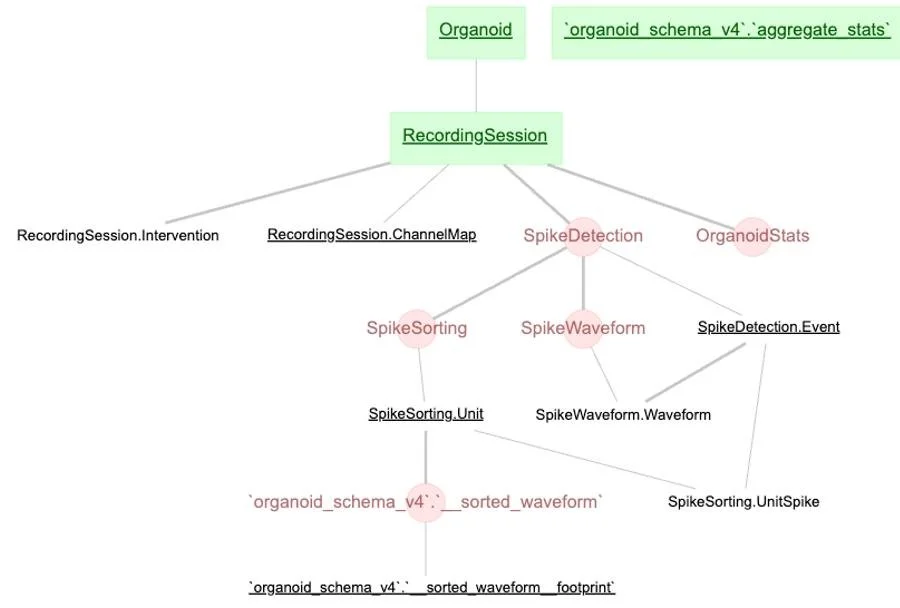
Visualization of Database Schema
Signal processing & spike sorting – Integrated open-source spike detection and sorting tools (SpikeInterface, Kilosort, MountainSort4) into a reproducible pipeline for our specific project, including filtering, artifact rejection, unit stability metrics, and waveform-based feature extraction for downstream modeling.
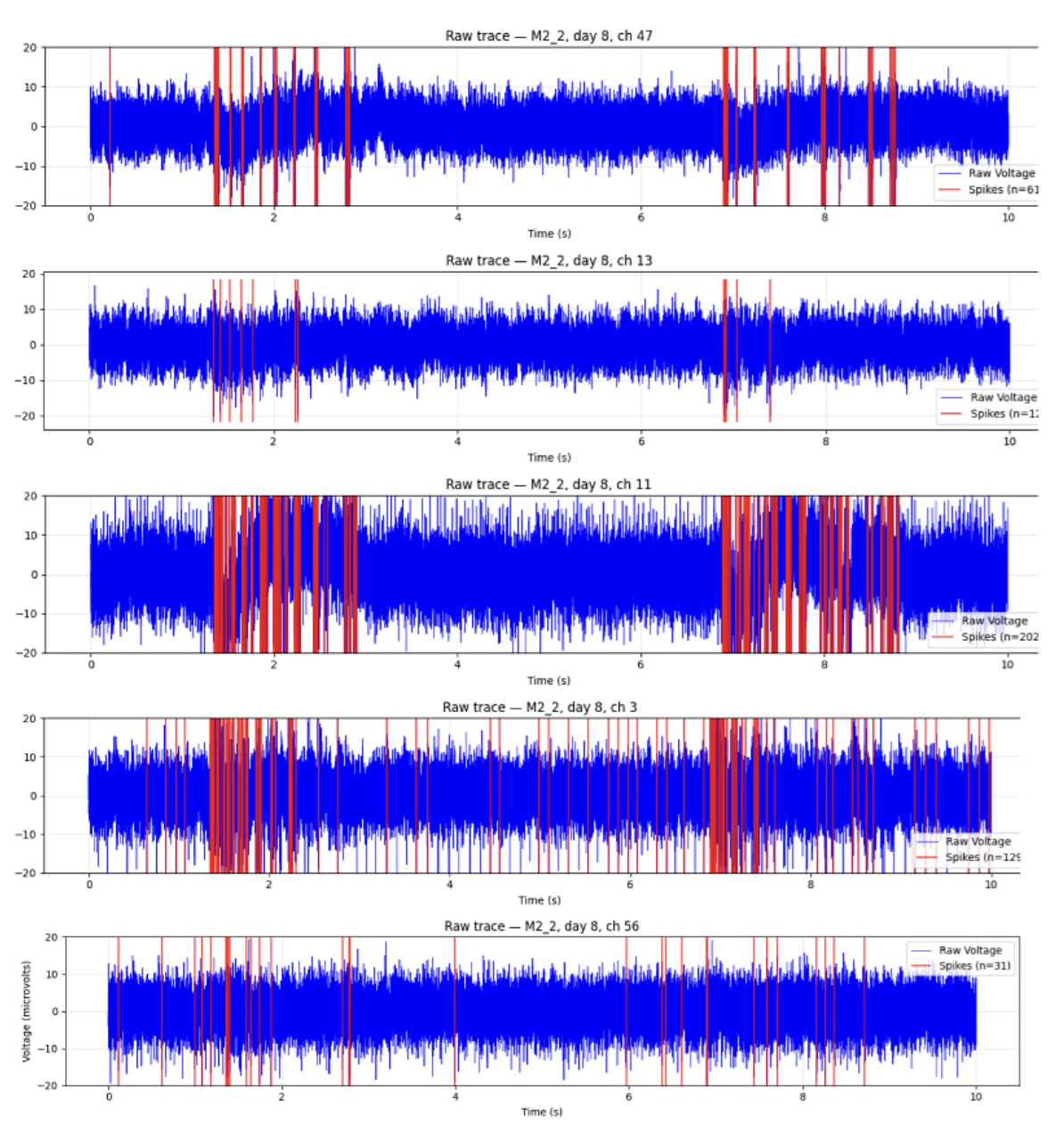
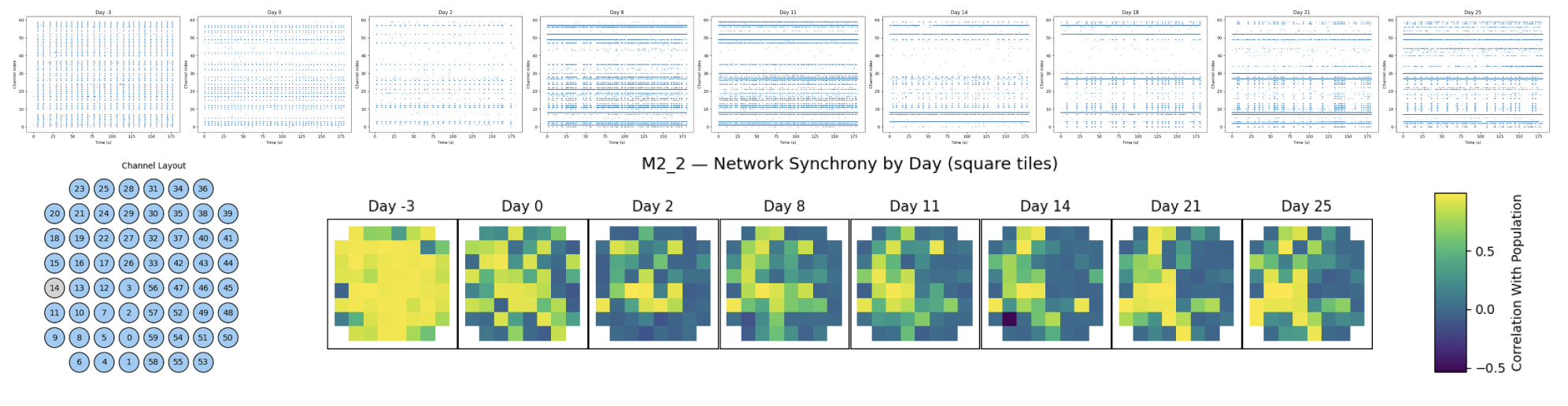
Channel Correlated Neural Bursting Activity
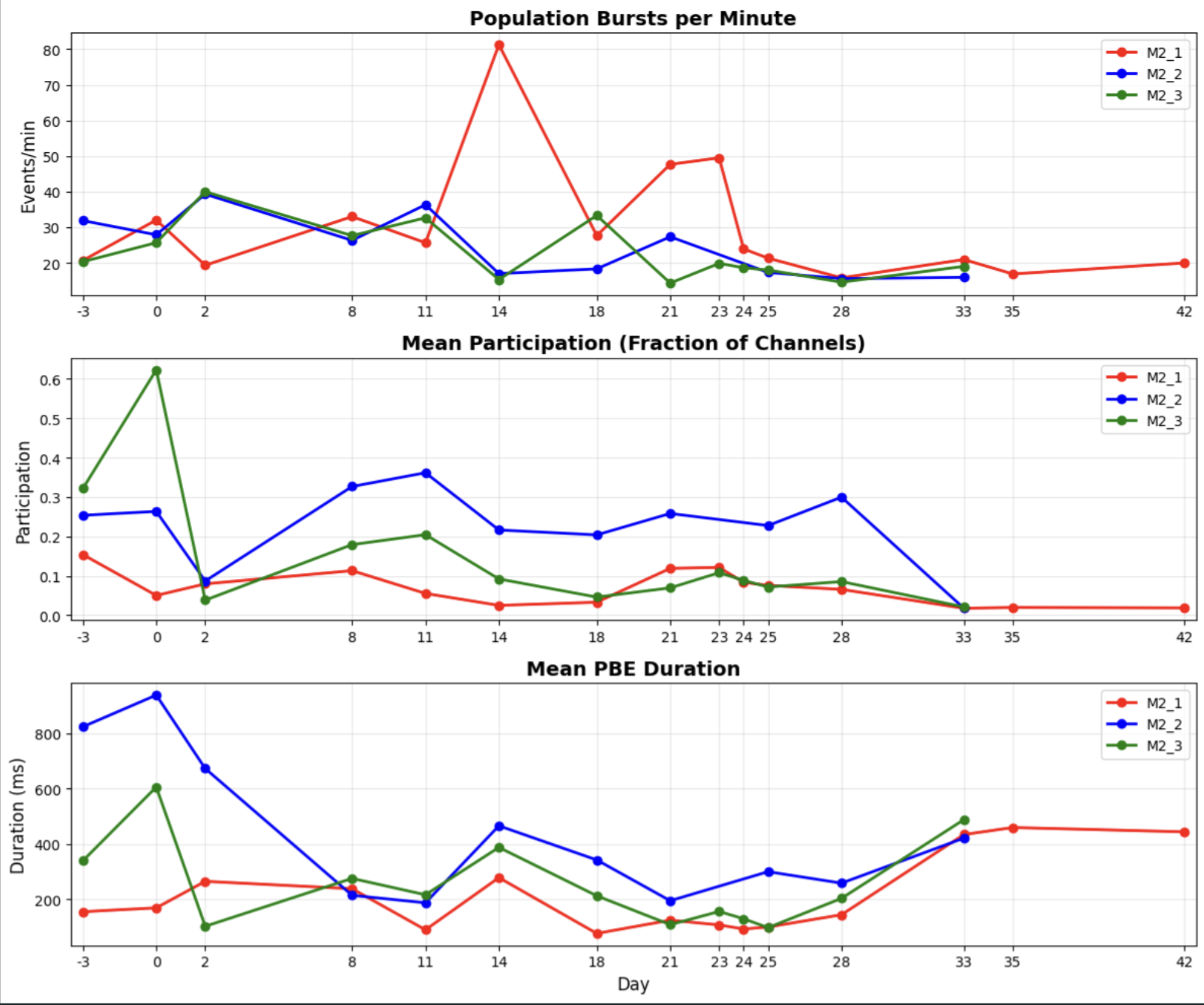
Quality control & network-level metrics – Defined quantitative QC metrics (firing rates, ISI violations, bursting, noise signatures, electrode stability) and built visualizations (e.g. heatmaps of neural activity organized by electrode geometry), computed network statistics to understand population activity and indentify problematic channels that might need to be excluded
Visualizing Network Development and Spiking Synchronicity Across Days
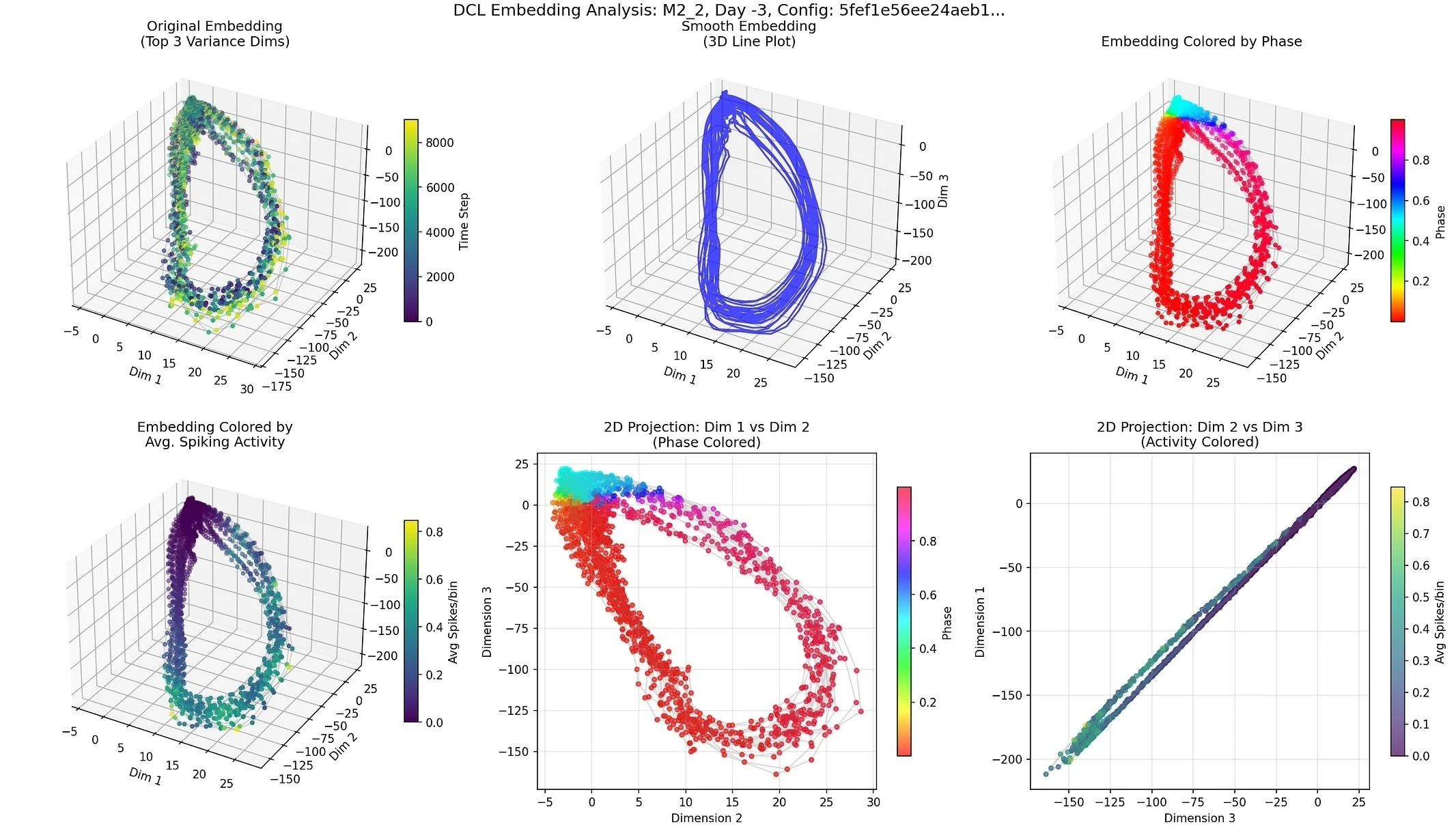
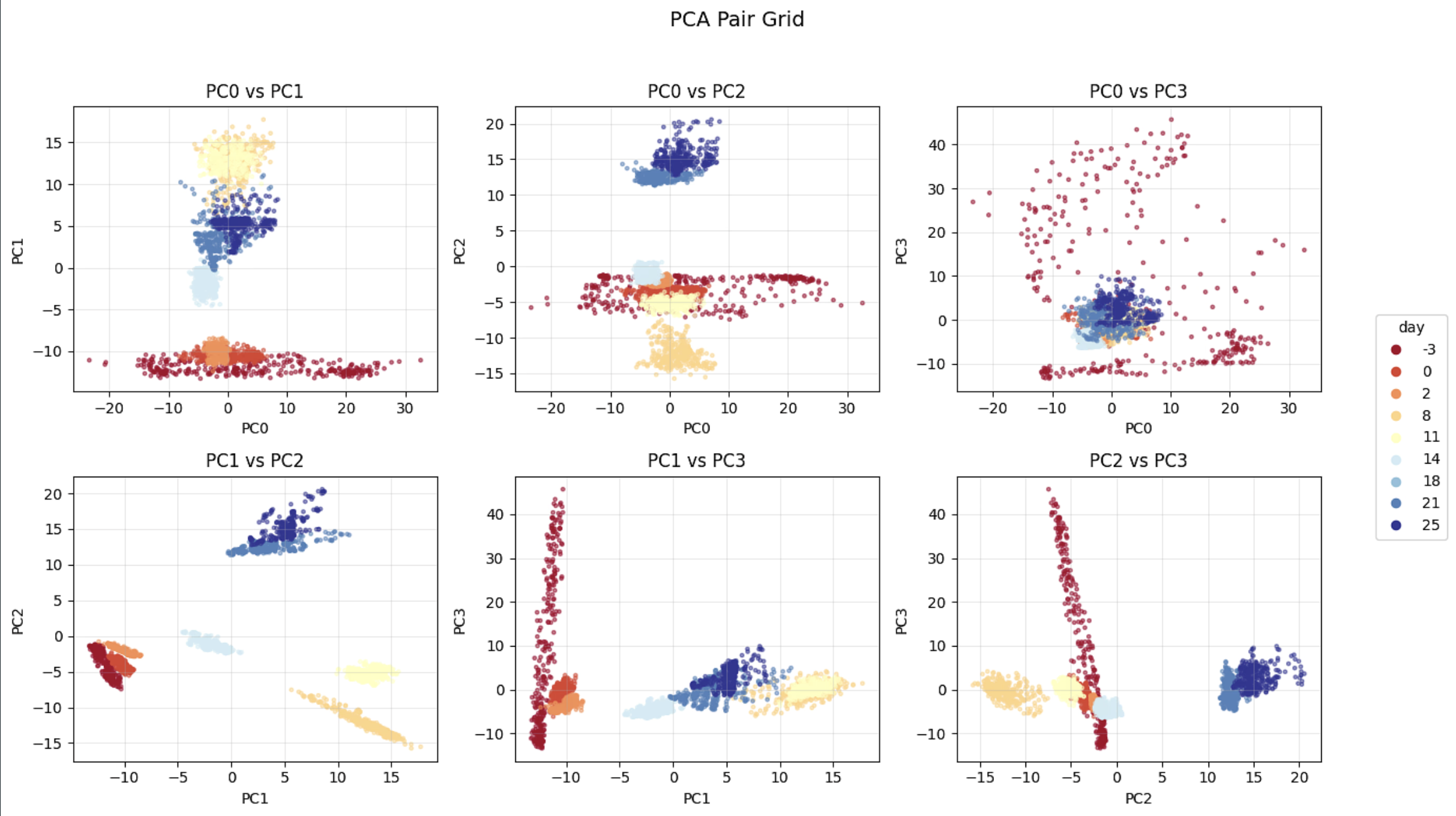
DCL Embedding Reveals a Smooth Cyclic Trajectory in Neural Activity
Example of Table from Schema (metadata and stats data)
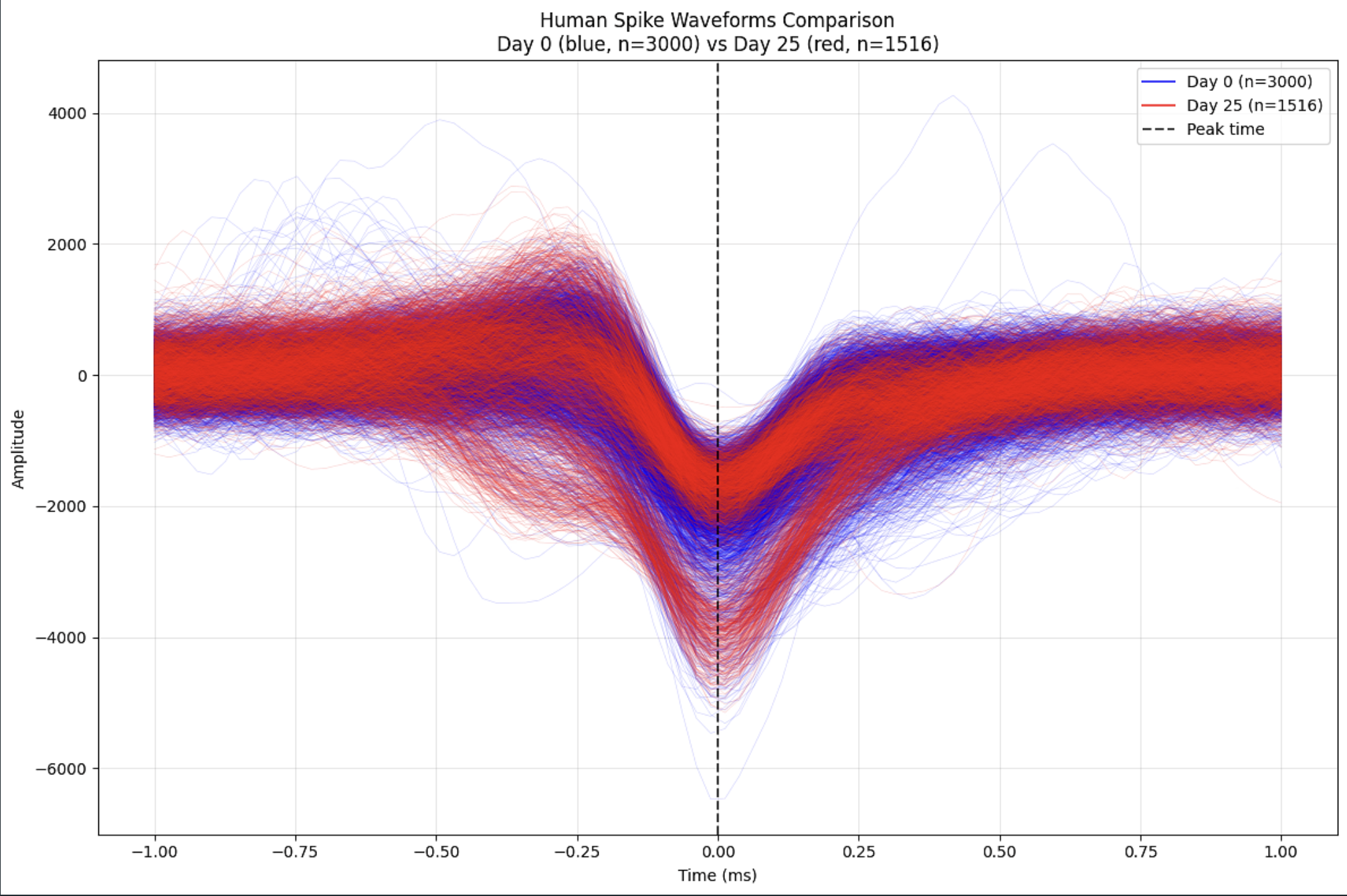
Comparison of Spike Waveform Shape After 25 Days of Development
Network Statistics Across Different Brain Organoids and Time
Machine learning & dimensionality reduction for neural dynamics – Modeled organoid activity as a dynamical system using time-delay embeddings; trained Dynamics Contrastive Learning (DCL) and related models to learn low-dimensional network-state spaces; and used PCA, UMAP, and regression models to interpret these spaces and compare spontaneous vs. evoked dynamics over development.
Pairwise PCA Projections Show Day-Specific Clustering of Neural Activity
Reproducible ML experiments – Structured and databased model configs, training runs, checkpoints, and embeddings so every latent space is traceable back to specific recordings and preprocessing choices, making complex analyses repeatable and auditable
HPC & robust pipelines – Set up and maintained analysis workflows across laptop, cloud virtual machines, and and high performance computing clusters; synced multi-TB datasets across systems and computed checksums to ensure the integrity and validity of data transfers; and configured long-running jobs with fail-safes so ML and preprocessing pipelines run reliably without manual babysitting
Time-series Integration across Modalities for Evaluation of Latent Dynamics
Motivation for Project
My Role:
Engineered data from 30,000 subjects that lead directly to success grant application.
Responsible for behavioral trajectory data engineering: transforming irregular raw tracking into standardized, model-ready time series at scale.
Drove the modeling workflow for representation learning on open-field behavior (CEBRA), including the shift to CEBRA-discrete and careful control over how training pairs/labels are formed.
Led metadata wrangling + integration logic for Open Field worksheets, building reliable filtering and year-extraction rules to support reproducible analysis.
Acted as the bridge between raw data realities (messy sampling, file structure, multi-mouse/multi-session organization) and clean ML training inputs that your lab can iterate on.
Other Credits: Steffen Schneider, Gil Westmeyer, Carsten Marr, Sabine Hölter-Koch, Christian Müller, Malte Lücken, German Mouse Clinic, Stanford University
Published Information: click here
Project Status: in progress
Large-scale behavioral datasets are only as valuable as their structure, consistency, and interpretability. The German Mouse Clinic (GMC) produces one of the most comprehensive phenotyping datasets available, but its scale and heterogeneity make downstream analysis and benchmarking challenging. The motivation in this project is to transform complex, multi-year behavioral data into a rigorously curated, well-documented resource that could support reproducible machine-learning research and cross-study comparison, rather than remaining locked in ad-hoc analysis pipelines. This work directly contributed to a successful Helmholtz Imaging grant application (TIMELY), and the curated dataset will serve as part of a public benchmarking suite scheduled for release next year, extending its impact beyond a single lab or project.
Technical Contribution
My contribution focused on turning raw German Mouse Clinic (GMC) behavioral outputs into a dataset that could actually function inside a standardized benchmark like TIMELY—meaning it had to be consistent, machine-learning–ready, and traceable back to the original source files.
Ingested and organized multi-year (30,000+ mice) GMC Open Field data and the accompanying metadata spreadsheets into a unified, structured dataset.
Designed automated database to standardized identifiers and joins (e.g., mouse IDs, line/genotype labels, cohort/session structure) so behavioral records reliably link to the correct metadata across files and years.
Cleaned and validated the dataset by checking for missing fields, inconsistent naming conventions, duplicate entries, and mismatched joins (and documenting these edge cases so they don’t silently propagate).
Converted raw trajectory/time-series outputs into analysis-ready formats, including reshaping data into consistent arrays/tables suitable for downstream modeling.
Applied machine learning methods to high-dimensional behavioral time-series data, preparing curated GMC datasets for use in embedding-based models, dynamical systems analyses, and standardized benchmarking tasks within the TIMELY framework.
Handled irregular time sampling by implementing preprocessing steps that support time-series models (e.g., interpolation/resampling decisions where needed) while preserving biological interpretability.
Prepared benchmark-friendly data artifacts (standard splits/exports and clear data dictionaries/field definitions) so the dataset can plug into shared evaluation tasks and reference code.
The result is a curated GMC dataset that is reproducible and extensible—exactly the type of “real biology” data TIMELY is designed to include—supporting benchmarking of dynamical models on complex, noisy behavioral time series.